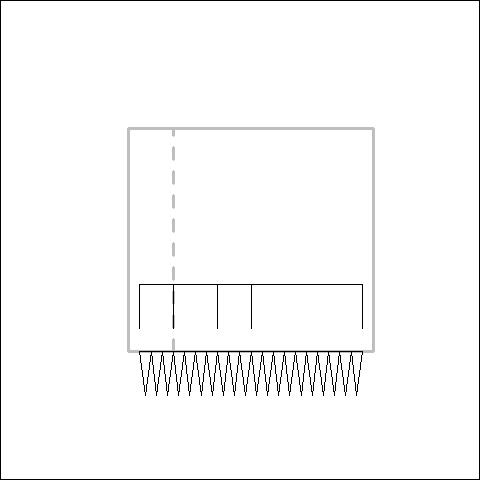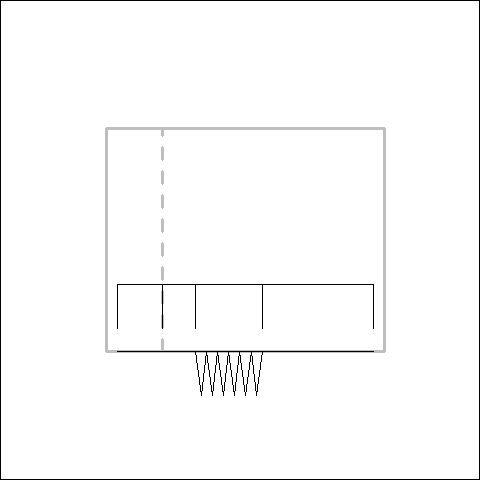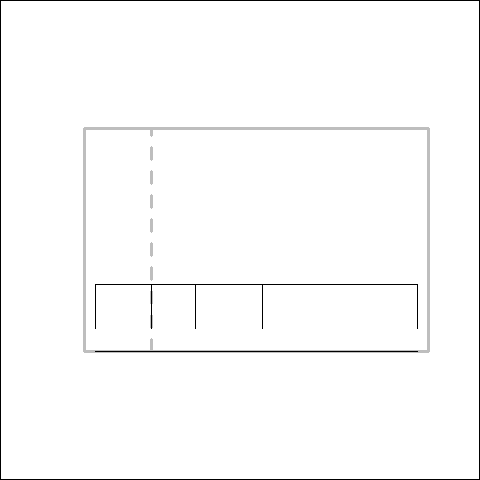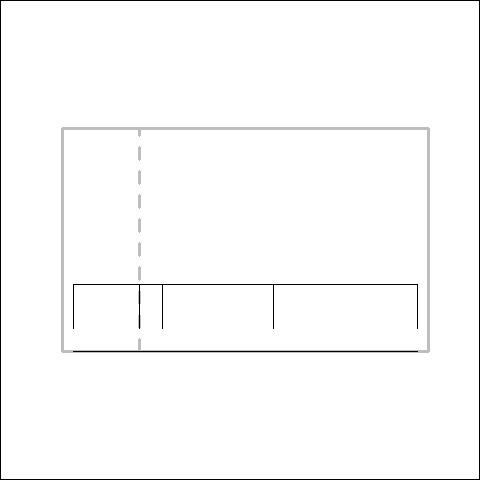
Simulating the Evolution of a Tooth Row
This method was described in Brocklehurst, N., Haridy, Y. 2020. Do meristic characters used in phylogenetic analysis evolve in an ordered manner? Systematic Biology (In Press). See this paper for references and more detail on the concepts
Evolution is complicated. I know there are a lot of people who think it isn't, and who decide that all it takes is 20 minutes on Google to become an expert. But it is actually really complicated. And sometimes even the people who study it forget just how complicated it can be.
Take, for example, ordered characters in phylogenetic analysis. For those of you unfamiliar, most commonly used methods of deciding evolutionary relationships using morphology work by tracking changes in discrete characters over a set of trees. Fewer changes mean a better tree (there are nuances and some more complex methods available, but they are underlain by these principles). The discrete characters used can be simple yes/no answers e.g. the astragalus (an ankle bone): absent or present? But in other cases they can be continuous characters that have just been discretised, like the number of teeth. And these bring their own sets of decisions to make, such as whether or not the character should be ordered
In phylogenetic analysis, most characters will be unordered: getting from character state 0 to state 1 is just as easy (and is counted as just as many changes) as getting to state 2. An ordered character requires a transition from state 0 to state 2 to go through state 1, taking two transitions. For example, to get from four teeth to six teeth you have to go through five teeth. And for a character like number of teeth you can see the logic behind this. Surely, to get from four teeth to six is harder than to five, and our methods should reflect this! But remember: nothing is ever so simple in evolution.
The development of most traits is under the control of a genetic regulatory network that forms a hierarchy of control. Hit the hierarchy at a high enough level and larger changes can be make. Like, for example, supressing the development of the entire tooth row rather than knocking out each tooth one by one in an ordered manner. You can also within the tooth row have different regions evolving semi-independently from each other. Canines, incisors, molars, all will be under differing selection pressures, and may change size and be supressed independently. To what extent will this affect whether the trait actually evolved in an ordered manner?
In order to answer this question, myself and Yara Haridy designed a simulation to see how these factors affect whether a tooth row evolves in an ordered manner or not. The base line of the simulation is a tooth row evolving by adding or subtracting one tooth at a time. Then we have some modularity. The jaw is split into four regions which change size through time independently of the changing number of teeth. Finally we have our regulatory hierarchy: a set of on/off characters that supress the development of the teeth in single regions, single bones or the entire tooth row.
Evolution is complicated. I know there are a lot of people who think it isn't, and who decide that all it takes is 20 minutes on Google to become an expert. But it is actually really complicated. And sometimes even the people who study it forget just how complicated it can be.
Take, for example, ordered characters in phylogenetic analysis. For those of you unfamiliar, most commonly used methods of deciding evolutionary relationships using morphology work by tracking changes in discrete characters over a set of trees. Fewer changes mean a better tree (there are nuances and some more complex methods available, but they are underlain by these principles). The discrete characters used can be simple yes/no answers e.g. the astragalus (an ankle bone): absent or present? But in other cases they can be continuous characters that have just been discretised, like the number of teeth. And these bring their own sets of decisions to make, such as whether or not the character should be ordered
In phylogenetic analysis, most characters will be unordered: getting from character state 0 to state 1 is just as easy (and is counted as just as many changes) as getting to state 2. An ordered character requires a transition from state 0 to state 2 to go through state 1, taking two transitions. For example, to get from four teeth to six teeth you have to go through five teeth. And for a character like number of teeth you can see the logic behind this. Surely, to get from four teeth to six is harder than to five, and our methods should reflect this! But remember: nothing is ever so simple in evolution.
The development of most traits is under the control of a genetic regulatory network that forms a hierarchy of control. Hit the hierarchy at a high enough level and larger changes can be make. Like, for example, supressing the development of the entire tooth row rather than knocking out each tooth one by one in an ordered manner. You can also within the tooth row have different regions evolving semi-independently from each other. Canines, incisors, molars, all will be under differing selection pressures, and may change size and be supressed independently. To what extent will this affect whether the trait actually evolved in an ordered manner?
In order to answer this question, myself and Yara Haridy designed a simulation to see how these factors affect whether a tooth row evolves in an ordered manner or not. The base line of the simulation is a tooth row evolving by adding or subtracting one tooth at a time. Then we have some modularity. The jaw is split into four regions which change size through time independently of the changing number of teeth. Finally we have our regulatory hierarchy: a set of on/off characters that supress the development of the teeth in single regions, single bones or the entire tooth row.
The simulation has been wrapped up in an R function that I'm going to show you all how to use now. Links to download the functions are in the tutorial
1. If not yet installed, install the R package phytools
2. Read in the phytools package
library(phytools)
3. Download and read in the tooth.sim() function (download via the link)
4. Generate a random tree by a birth death process (may need to rerun this line a couple of times; sometimes, if extinction happens too soon, a tree with only 1 or a small number of taxa is produced)
tree<-pbtree(b=0.1,d=0.05,n=100,scale=100)
5. Choose analysis parameters. The tooth.sim() function has seven arguments:
· tree: object of class phylo; the phylogeny over which evolution of the character is simulated. Required
· max.teeth: class numeric; the maximum number of teeth allowed. Required. Default 40
· param.H.1: class numeric; instantaneous transition rate of level 1 of the developmental controls: toothlessness or not. Can be NULL if such transitions are not wanted. Default 0.002
· param.H.2: class numeric; instantaneous transition rate of level 2 of the developmental controls: presence or absence of teeth in each bone (Maxilla or premaxilla). Can be NULL if such transitions are not wanted. Default 0.002
· param.H.3: class numeric; instantaneous transition rate of level 3 of the developmental controls: presence or absence of teeth in each dental region (incisor, caniniform, premolar or molar). Can be NULL if such transitions are not wanted. Default 0.002
· param.T: class numeric; instantaneous transition rate of level 4 of the developmental controls: addition or subtraction of teeth one by one. Required. Default 0.002
· param.R: class numeric; sigma2 (i.e. rate) parameter of the brownian motion models used to simulate changes in relative size of the regions (incisor, caniniform, premolar or molar). Can be NULL; if so the tooth row will evolve as a single unit with no regionality. Default 0.04
7. Run a simulation. This example does not include any modularity. The object produced is a vector giving the number of teeth of each taxon
test<-tooth.sim(tree,max.teeth=40,param.H.1=NULL,param.H.2=NULL,param.H.3=NULL, param.T=0.5,param.R=NULL)
test
8. Now run a second simulation, this time with regionality of the tooth row. The object produced this time is a matrix, giving for each taxon (rows) the number of teeth in each tooth region (columns).
test2<-tooth.sim(tree,max.teeth=40,param.H.1=0.002,param.H.2=0.002,param.H.3=0.002, param.T=0.5,param.R=0.04)
test2
1. If not yet installed, install the R package phytools
2. Read in the phytools package
library(phytools)
3. Download and read in the tooth.sim() function (download via the link)
4. Generate a random tree by a birth death process (may need to rerun this line a couple of times; sometimes, if extinction happens too soon, a tree with only 1 or a small number of taxa is produced)
tree<-pbtree(b=0.1,d=0.05,n=100,scale=100)
5. Choose analysis parameters. The tooth.sim() function has seven arguments:
· tree: object of class phylo; the phylogeny over which evolution of the character is simulated. Required
· max.teeth: class numeric; the maximum number of teeth allowed. Required. Default 40
· param.H.1: class numeric; instantaneous transition rate of level 1 of the developmental controls: toothlessness or not. Can be NULL if such transitions are not wanted. Default 0.002
· param.H.2: class numeric; instantaneous transition rate of level 2 of the developmental controls: presence or absence of teeth in each bone (Maxilla or premaxilla). Can be NULL if such transitions are not wanted. Default 0.002
· param.H.3: class numeric; instantaneous transition rate of level 3 of the developmental controls: presence or absence of teeth in each dental region (incisor, caniniform, premolar or molar). Can be NULL if such transitions are not wanted. Default 0.002
· param.T: class numeric; instantaneous transition rate of level 4 of the developmental controls: addition or subtraction of teeth one by one. Required. Default 0.002
· param.R: class numeric; sigma2 (i.e. rate) parameter of the brownian motion models used to simulate changes in relative size of the regions (incisor, caniniform, premolar or molar). Can be NULL; if so the tooth row will evolve as a single unit with no regionality. Default 0.04
7. Run a simulation. This example does not include any modularity. The object produced is a vector giving the number of teeth of each taxon
test<-tooth.sim(tree,max.teeth=40,param.H.1=NULL,param.H.2=NULL,param.H.3=NULL, param.T=0.5,param.R=NULL)
test
8. Now run a second simulation, this time with regionality of the tooth row. The object produced this time is a matrix, giving for each taxon (rows) the number of teeth in each tooth region (columns).
test2<-tooth.sim(tree,max.teeth=40,param.H.1=0.002,param.H.2=0.002,param.H.3=0.002, param.T=0.5,param.R=0.04)
test2